Post by Mikhail Fursov on Sep 7th, 2011 at 8:36pm
This is looks strange to me, I use the same version and OS and alignment with MUSCLE does work.
Lets check this with the file in attachment using the following scenario
1) open this file as alignment
2) insert space before the C letter
3) run alignment with muscle algorithm
The file must be realigned to initial state before the space is inserted and file name in the project view must be in dark blue color, but not black.
Could you check this scenario please?
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=test.fa (0 KB | )
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=test.fa (0 KB | )
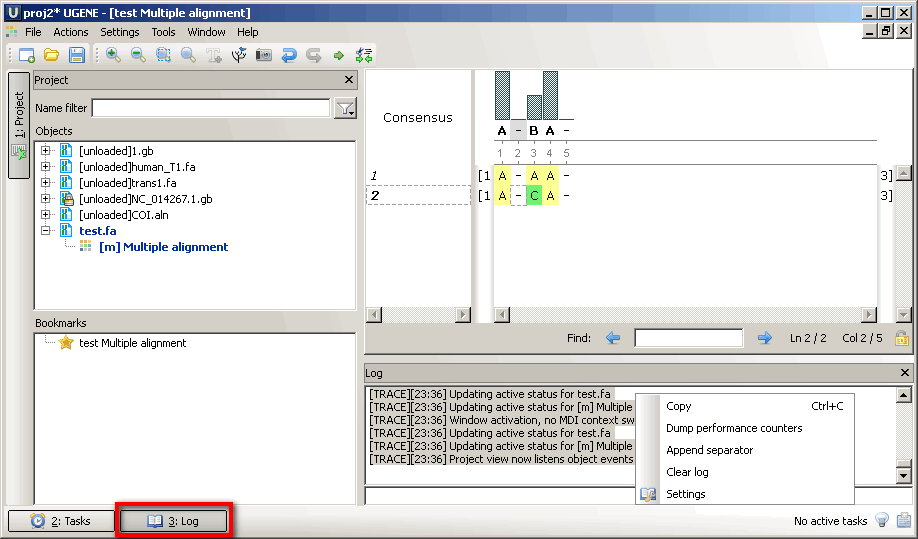
Lets check this with the file in attachment using the following scenario
1) open this file as alignment
2) insert space before the C letter
3) run alignment with muscle algorithm
The file must be realigned to initial state before the space is inserted and file name in the project view must be in dark blue color, but not black.
Could you check this scenario please?

 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=log_ugene_test.txt (68 KB | )
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=log_ugene_test.txt (68 KB | )