Post by Mikhail Fursov on Mar 13th, 2009 at 6:15pm
Method 1
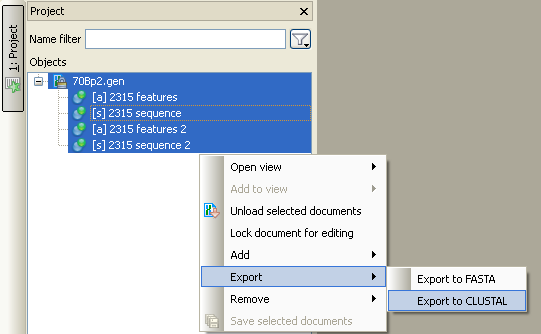
Step 1: Open FASTA (or any other file with sequences) to the active project
Step 2: Select the sequence objects you want to add to the alignment. Note that if you select the document with sequence objects, all sequences from the document will be added
Step 3: Use "Export->Export to CLUSTAL" menu.
After you click this menu you'll be proposed to select an alignment file to save results to.
 1_002.png (9 KB | )
1_002.png (9 KB | )
Step 1: Open FASTA (or any other file with sequences) to the active project
Step 2: Select the sequence objects you want to add to the alignment. Note that if you select the document with sequence objects, all sequences from the document will be added
Step 3: Use "Export->Export to CLUSTAL" menu.
After you click this menu you'll be proposed to select an alignment file to save results to.
 1_002.png (9 KB | )
1_002.png (9 KB | )