Post by R Smith on May 31st, 2012 at 2:53pm
Konstantin Okonechnikov wrote on May 31st, 2012 at 4:58am:
|
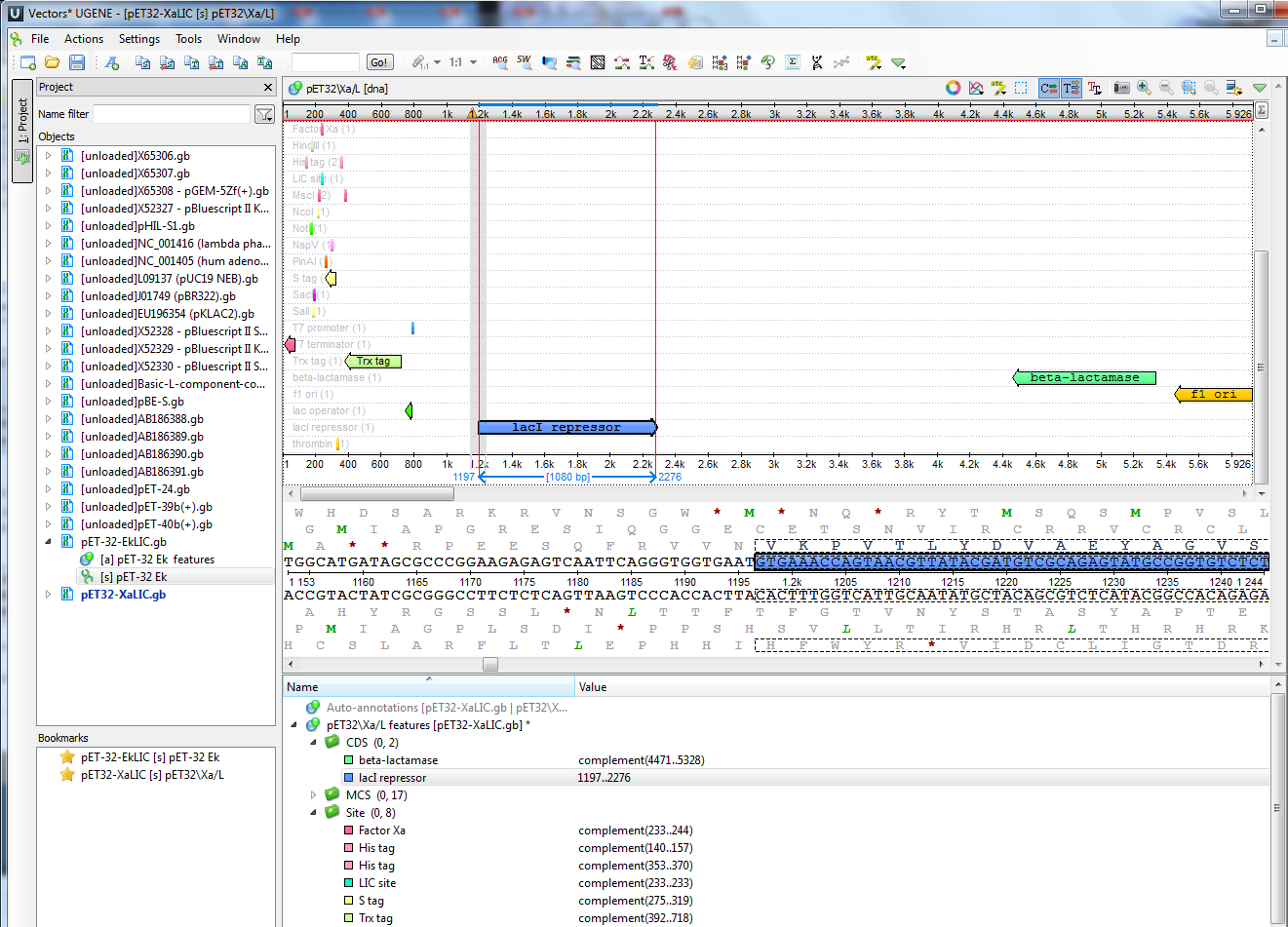
I have attached an image showing what I am talking about. It is in the sequence overview panel or subwindow at the top. The current interface shows light gray or silver-colored solid lines that delimit the selected annotation, along with the bp length and the delimiting nucleotide positions in light blue. The interface also has the movable sequence detail position indicator, with dashed very light gray lines showing which part of the overall sequence is detailed.
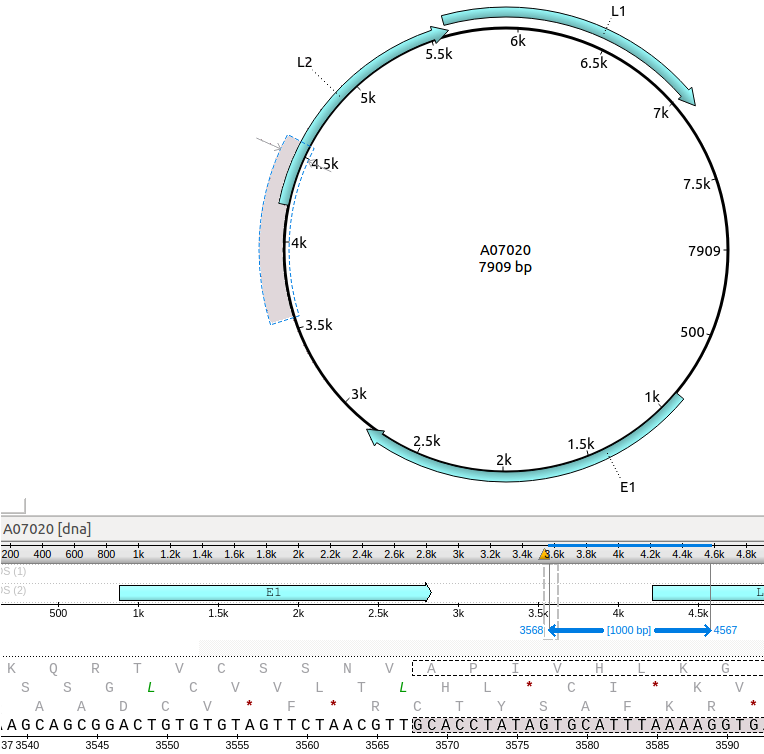
I modified both of these. I played around with a few styles of presentation and came up with what is imaged below. For the movable sequence detail indicator, I thought a solid light gray rectangle with the width spanning the presented sequence detail. For the elements (annotations, scale numbers, horizontal lines) with the sequence overiew, these elements should always be placed in front of the shaded rectangle.
As for the solid vertical lines highlighting the selected annotation, I went with a different color other than a silver-colored solid line.
It's just my opinion, but I think that light gray- and silver-colored thin lines against a white background doesn't provide eye-catching contrast for the user.
 ugene-linear-molecule-view-colored.png (233 KB | )
ugene-linear-molecule-view-colored.png (233 KB | )