Post by Yuriy Vaskin on Jan 16th, 2013 at 2:49pm
Hello!
To do that you need to use Workflow Designer.
I've attached a scheme for that task. I'm going to put it to sample schemes of UGENE so anyone can use it.
Just open the attached scheme and set up the following parameters:
1. In Read Sequence element select the input file(s) - your protein sequences.
2. In Find Substring element set patterns ( semicolon separated list of patterns) - your motifs.
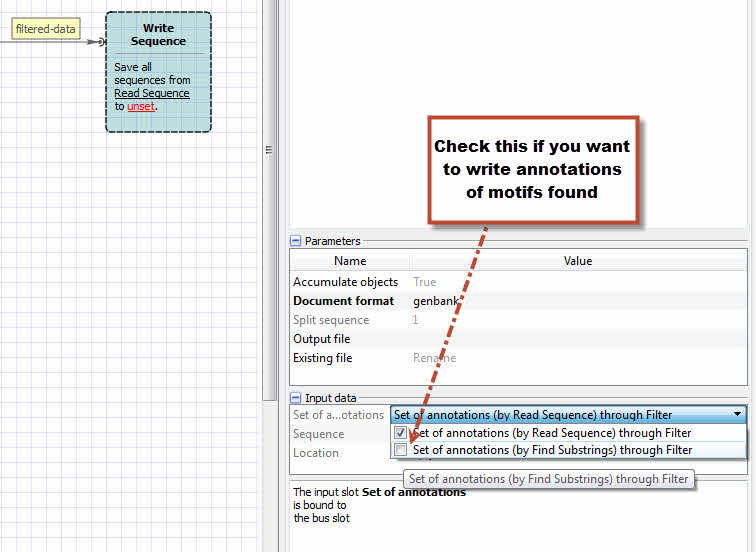
3. In Write Sequence element set the output file.
That scheme reads all your sequences, finds listed pattern in them and mark those sequences that have a motif of interest (number of annotations with name motif_of_interest >=1 ), it passes to the output only the sequences that has a motif.
You may play with the parameters of the elements of the scheme to see what will happen. These chapters of documentation might help: http://ugene.unipro.ru/documentation/wd_manual/index.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/data_flow/marker.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/data_flow/marker.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/basic_analysis/find_substrings.html
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=filter_by_motif.uwl (8 KB | )
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=filter_by_motif.uwl (8 KB | )
 motifs.jpg (63 KB | )
motifs.jpg (63 KB | )
To do that you need to use Workflow Designer.
I've attached a scheme for that task. I'm going to put it to sample schemes of UGENE so anyone can use it.
Just open the attached scheme and set up the following parameters:
1. In Read Sequence element select the input file(s) - your protein sequences.
2. In Find Substring element set patterns ( semicolon separated list of patterns) - your motifs.
3. In Write Sequence element set the output file.
That scheme reads all your sequences, finds listed pattern in them and mark those sequences that have a motif of interest (number of annotations with name motif_of_interest >=1 ), it passes to the output only the sequences that has a motif.
You may play with the parameters of the elements of the scheme to see what will happen. These chapters of documentation might help: http://ugene.unipro.ru/documentation/wd_manual/index.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/data_flow/marker.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/data_flow/marker.html
http://ugene.unipro.ru/documentation/wd_manual/workflow_elements/basic_analysis/find_substrings.html
 motifs.jpg (63 KB | )
motifs.jpg (63 KB | )