Post by Konstantin Okonechnikov on Aug 13th, 2010 at 6:29pm
Hello!
As I understand, you need to save several sequences into one file. What is the expected output format for the isoform map? Are the annoations required for the map? It would be great if you provide some details.
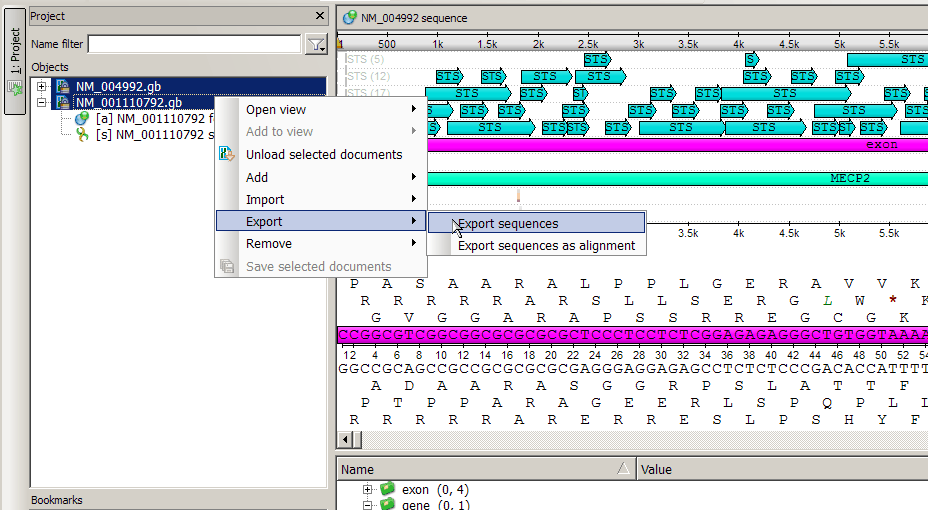
Anyway the easiest way is to use Ugene export function. Just open the required documents and use export context menu in the Project view.
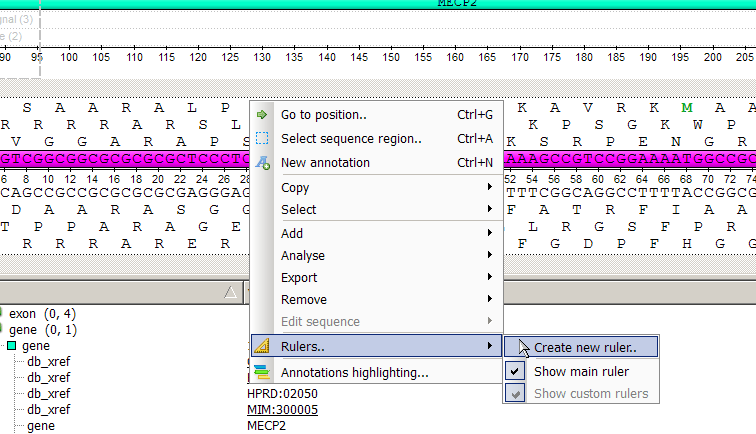
As for adding new ruler : in the sequence view activate context menu with right mouse button. Select "Rulers-> create new ruler"
In the appeared dialog you can set ruler name, starting position (offset) and color.
Check UGENE user manual for more details: http://ugene.unipro.ru/manual/adv_pan_view.html
 ScreenHunter_02_Aug__13_18_18.gif (39 KB | )
ScreenHunter_02_Aug__13_18_18.gif (39 KB | )
 ScreenHunter_04_Aug__13_18_27.gif (24 KB | )
ScreenHunter_04_Aug__13_18_27.gif (24 KB | )
As I understand, you need to save several sequences into one file. What is the expected output format for the isoform map? Are the annoations required for the map? It would be great if you provide some details.
Anyway the easiest way is to use Ugene export function. Just open the required documents and use export context menu in the Project view.
As for adding new ruler : in the sequence view activate context menu with right mouse button. Select "Rulers-> create new ruler"
In the appeared dialog you can set ruler name, starting position (offset) and color.
Check UGENE user manual for more details: http://ugene.unipro.ru/manual/adv_pan_view.html
 ScreenHunter_02_Aug__13_18_18.gif (39 KB | )
ScreenHunter_02_Aug__13_18_18.gif (39 KB | ) ScreenHunter_04_Aug__13_18_27.gif (24 KB | )
ScreenHunter_04_Aug__13_18_27.gif (24 KB | )