Post by Dmitrii Sukhomlinov on Dec 13th, 2021 at 11:55am
Hello,
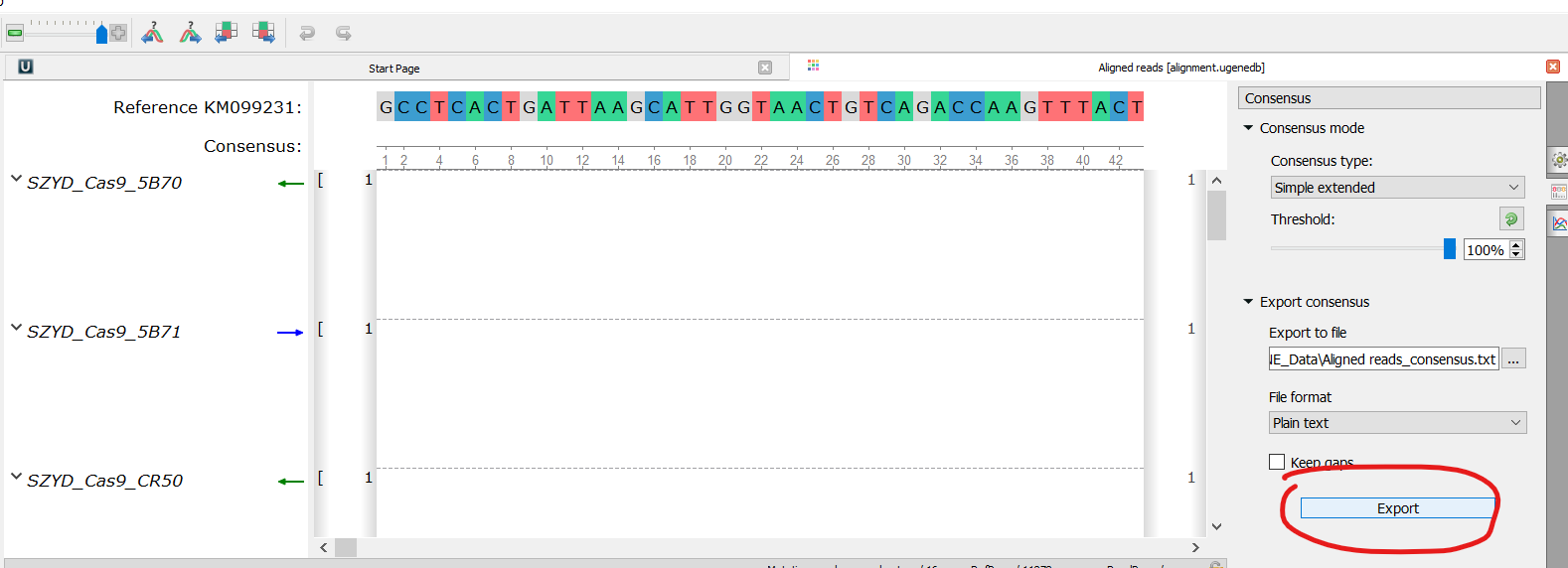
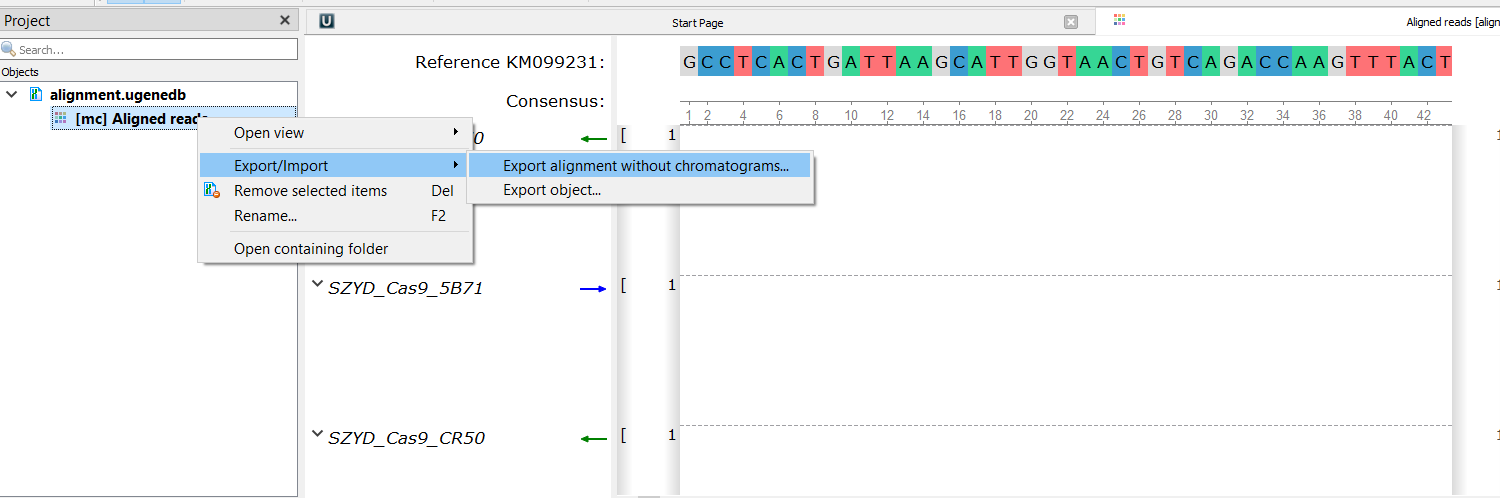
Could you, please, detail this a bit. How this feature differs from the consensus? You may export the consensus from the Sanger Reads Editor as a sequence (see attachment 1), also, you may export the whole Sanger alignment as Multiple alignment with the consensus (see attachment 2). Isn't this fit to you?
 1_012.png (48 KB | 201
)
1_012.png (48 KB | 201
)
 2_010.png (33 KB | 181
)
2_010.png (33 KB | 181
)
Could you, please, detail this a bit. How this feature differs from the consensus? You may export the consensus from the Sanger Reads Editor as a sequence (see attachment 1), also, you may export the whole Sanger alignment as Multiple alignment with the consensus (see attachment 2). Isn't this fit to you?
 1_012.png (48 KB | 201
)
1_012.png (48 KB | 201
) 2_010.png (33 KB | 181
)
2_010.png (33 KB | 181
)