Post by Dmitrii Sukhomlinov on Jan 22nd, 2021 at 3:33pm
Hello,
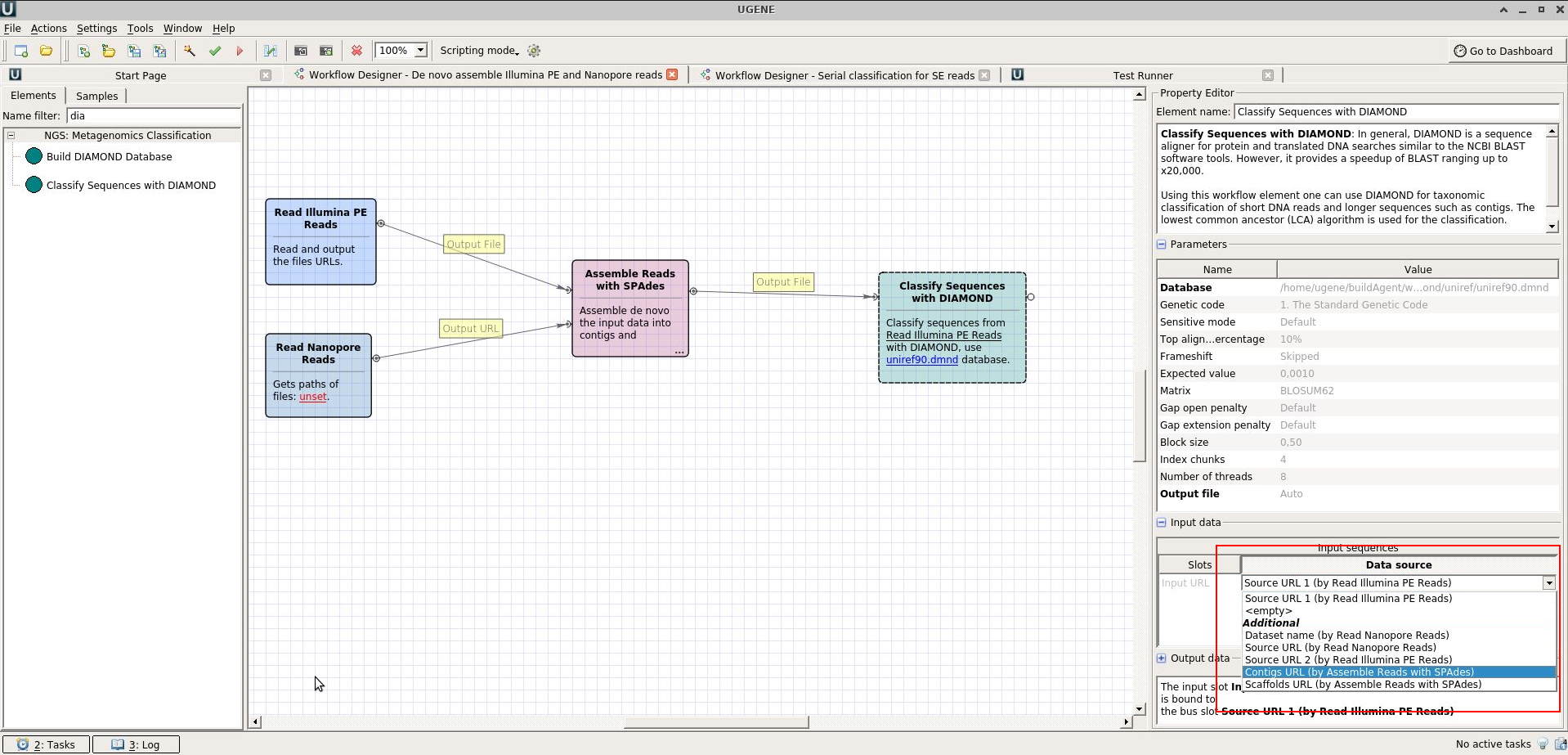
First of all, make sure, that the "DIAMOND" element input port is set to the correct output of SPAdes (see the attached picture).
Also, make sure, that you use the last UGENE v. 37 (there were some problems with diamond which were fixed).
If both points are OK, please, share your data with me if it's possible and I will try to reproduce the problem.
 Screenshot_1_001.jpg (241 KB | 277
)
Screenshot_1_001.jpg (241 KB | 277
)
First of all, make sure, that the "DIAMOND" element input port is set to the correct output of SPAdes (see the attached picture).
Also, make sure, that you use the last UGENE v. 37 (there were some problems with diamond which were fixed).
If both points are OK, please, share your data with me if it's possible and I will try to reproduce the problem.
 Screenshot_1_001.jpg (241 KB | 277
)
Screenshot_1_001.jpg (241 KB | 277
)