Post by Nikolas on May 22nd, 2019 at 5:16pm
Hello Olga,
thanks for your reply.
I have attached three of the read files to this post, the fourth I will attatch to the next post, as it is only possible to attatch three 250 KB files and each of them is 150 KB.
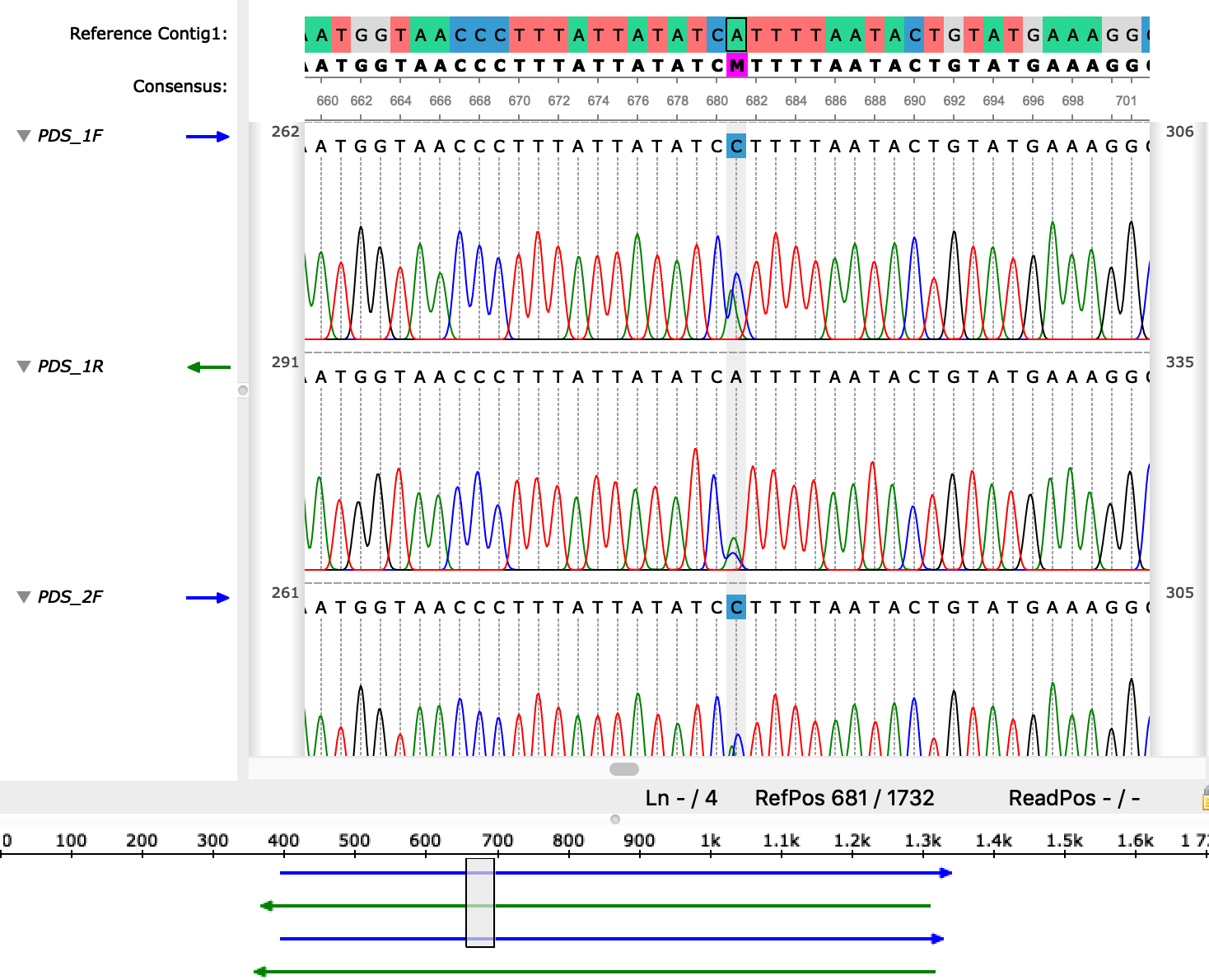
I did not have a reference sequence and my idea was to assemble the reads without one. Now I of course can use the consensus form the MUSCLE alignment as a reference for the reads. I have tried it as a workaraound and it works fine.
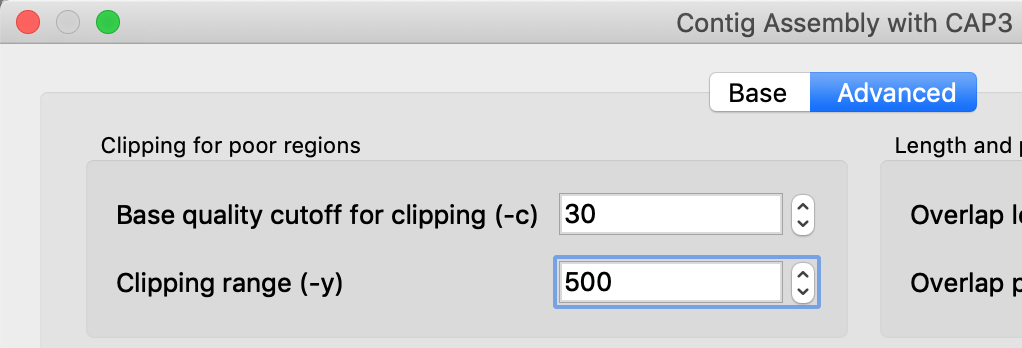
It would still be more convenient to directly assemble the reads with CAP3. Which parameters would I have to tweak? As far as I can see, two of my sequences are just being ignored and I don't see any parameters in the Advanced tab that would influence this behavior. Except maybe for the "Assembly reverse reads" flag which I have set on and off without any effect on the outcome.
Cheers,
Nikolas
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1F_ab1.zip (148 KB | 493
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1F_ab1.zip (148 KB | 493
) https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1R_ab1.zip (145 KB | 516
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1R_ab1.zip (145 KB | 516
) https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_2F_ab1.zip (146 KB | 322
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_2F_ab1.zip (146 KB | 322
)
thanks for your reply.
I have attached three of the read files to this post, the fourth I will attatch to the next post, as it is only possible to attatch three 250 KB files and each of them is 150 KB.
I did not have a reference sequence and my idea was to assemble the reads without one. Now I of course can use the consensus form the MUSCLE alignment as a reference for the reads. I have tried it as a workaraound and it works fine.
It would still be more convenient to directly assemble the reads with CAP3. Which parameters would I have to tweak? As far as I can see, two of my sequences are just being ignored and I don't see any parameters in the Advanced tab that would influence this behavior. Except maybe for the "Assembly reverse reads" flag which I have set on and off without any effect on the outcome.
Cheers,
Nikolas
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1F_ab1.zip (148 KB | 493
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1F_ab1.zip (148 KB | 493
) https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1R_ab1.zip (145 KB | 516
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_1R_ab1.zip (145 KB | 516
) https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_2F_ab1.zip (146 KB | 322
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=PDS_2F_ab1.zip (146 KB | 322
) https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=Contig1.fa (1 KB | 321
)
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=Contig1.fa (1 KB | 321
)