Post by AK on Apr 25th, 2014 at 3:56am
Hi,
I am trying your workflow designer for the first time and am really impressed by the progress Ugene has made in past few years. Nice work !!
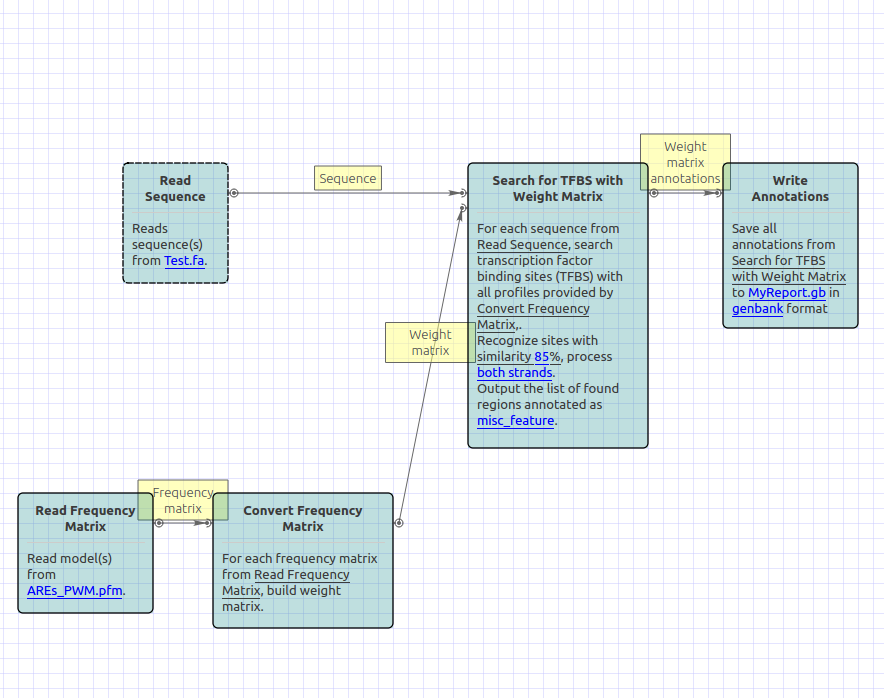
But I need your help in getting the results out in human readable format. So all I want to do is read a frequency matrix and search a set of sequences (n=46) for the TFBS. Finally output the results in a format that tells me the sequence name, coordinates of match,length,score etc.
To do so I created a workflow (attached) but there are two problems I am facing 1) The output format is 'annotation' and it shows result for only one sequence instead of all 46 sequences 2) 'Annotation' output format is not good for human reading, how can I get a csv format with sequence name, match co-ordinates and score. Is it possible to get a single result file for all 46 sequences?
Awaiting Reply
AK
 Selection_003.png (82 KB | )
Selection_003.png (82 KB | )
I am trying your workflow designer for the first time and am really impressed by the progress Ugene has made in past few years. Nice work !!
But I need your help in getting the results out in human readable format. So all I want to do is read a frequency matrix and search a set of sequences (n=46) for the TFBS. Finally output the results in a format that tells me the sequence name, coordinates of match,length,score etc.
To do so I created a workflow (attached) but there are two problems I am facing 1) The output format is 'annotation' and it shows result for only one sequence instead of all 46 sequences 2) 'Annotation' output format is not good for human reading, how can I get a csv format with sequence name, match co-ordinates and score. Is it possible to get a single result file for all 46 sequences?
Awaiting Reply
AK
 Selection_003.png (82 KB | )
Selection_003.png (82 KB | )