Post by Konstantin Okonechnikov on May 8th, 2012 at 4:11am
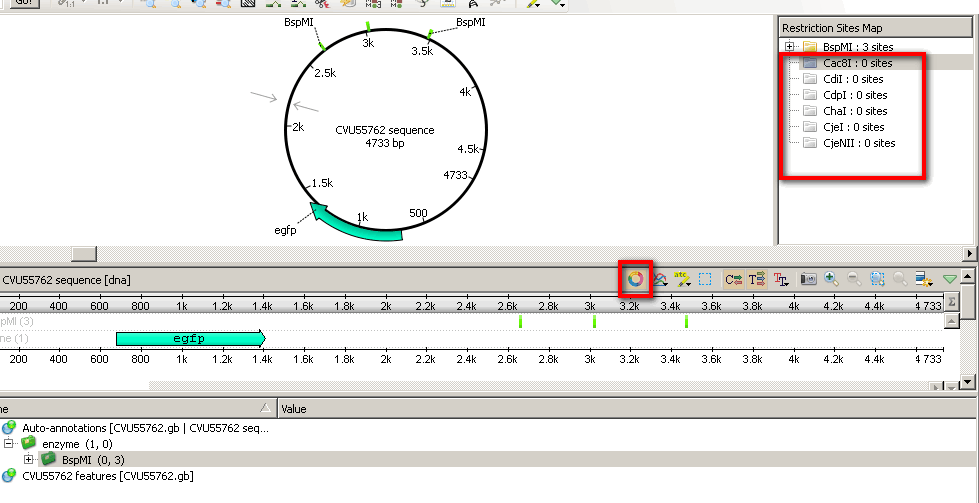
Hi! I am not sure if I get it right. Do you want to find all enzymes, which do not cut the sequence or do you want to know if some specific enzymes do not cut the sequence? In the latter case you can just select a group of enzymes and then use Restriction Sites Map from Circular View in order to see which restriction sites are not present. (see screenshot attached)
 screenshot_restrction_sites_map.gif (26 KB | )
screenshot_restrction_sites_map.gif (26 KB | )
 screenshot_restrction_sites_map.gif (26 KB | )
screenshot_restrction_sites_map.gif (26 KB | )