Post by Yuriy Vaskin on Apr 12th, 2012 at 6:08pm
Thanks for using UGENE!
Let's see if I understand your issue correctly.
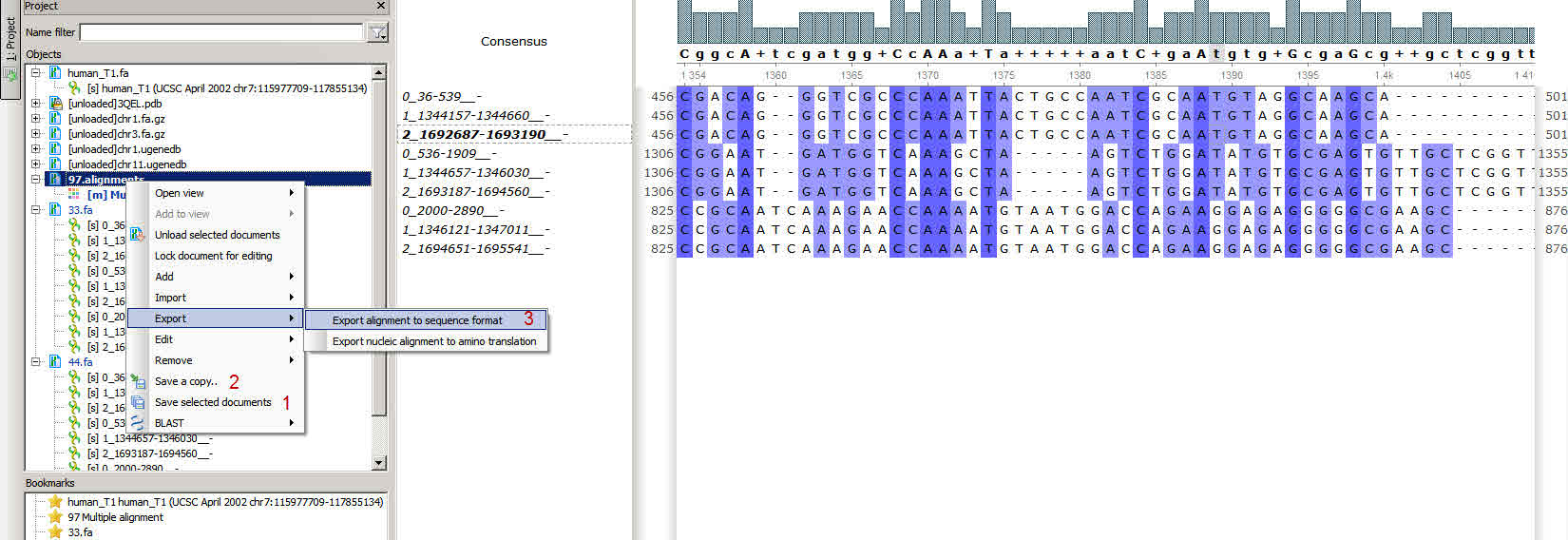
You open a multiple FASTA file, export it into alignment (have you done that using "Join sequences into alignment..." option in the "Sequence reading option" dialog when you open your file?), then align it. Your original file is not modified yet, unless you click on "Save selected document" (pic1 - 1). Then the original file is overwritten.
Just click on the "Save a copy" (pic1 - 2) to keep original file and save a modified one into another file. By the way, you may choose another format for the alignment. FASTA is not a good one for that. If you choose a multiple alignment format (CLUSTALW, Mega, ...) there will be no problem with opening it by other tools.
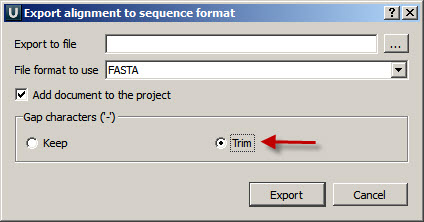
Also you may export your alignment into sequences (Right button on the document - Export - Export alignment to sequence format)(pic1 - 3). And remove dashes from the output file by choosing the corresponding option (pic 2.)
Please let me know if it doesn't work for you.
 al1.jpg (137 KB | )
al1.jpg (137 KB | )
 al2.jpg (28 KB | )
al2.jpg (28 KB | )
Let's see if I understand your issue correctly.
You open a multiple FASTA file, export it into alignment (have you done that using "Join sequences into alignment..." option in the "Sequence reading option" dialog when you open your file?), then align it. Your original file is not modified yet, unless you click on "Save selected document" (pic1 - 1). Then the original file is overwritten.
Just click on the "Save a copy" (pic1 - 2) to keep original file and save a modified one into another file. By the way, you may choose another format for the alignment. FASTA is not a good one for that. If you choose a multiple alignment format (CLUSTALW, Mega, ...) there will be no problem with opening it by other tools.
Also you may export your alignment into sequences (Right button on the document - Export - Export alignment to sequence format)(pic1 - 3). And remove dashes from the output file by choosing the corresponding option (pic 2.)
Please let me know if it doesn't work for you.
 al1.jpg (137 KB | )
al1.jpg (137 KB | ) al2.jpg (28 KB | )
al2.jpg (28 KB | )