Quote:Could you please describe a situation when this is option is not shown? What is the format of the file? Was it obtained from specific database or created by some software? If it is created by UGENE what was the pipeline?
|
|
Now I figure it out, so I can give a better explanation. My goal was, working with several genbank files in the same display, to select the same feature in the different sequences and to make an alignment. It would be nice to be able to do that for both, DNA and protein.
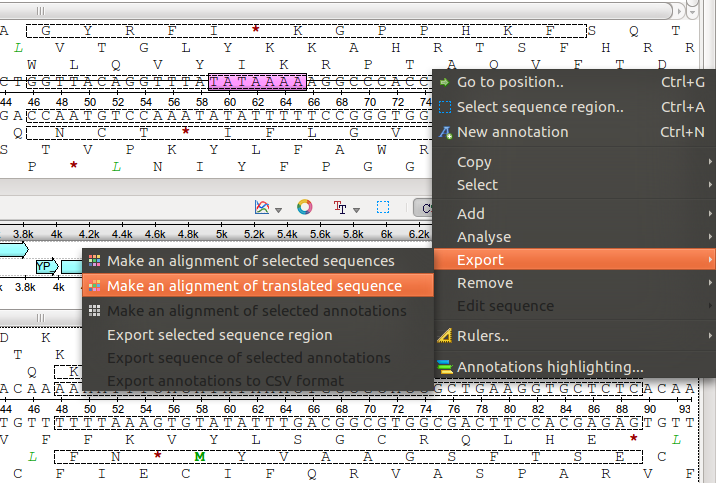
If I select the different features, I only have the option for a DNA alignment:
 http://ugene.unipro.ru/yabbfiles/Attachments/No_selection.png
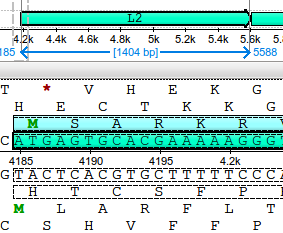
http://ugene.unipro.ru/yabbfiles/Attachments/No_selection.pngNow, if I have the Details view activated and select some sequence for both, I get the option for translated alignment:
 http://ugene.unipro.ru/yabbfiles/Attachments/Detail_view.png
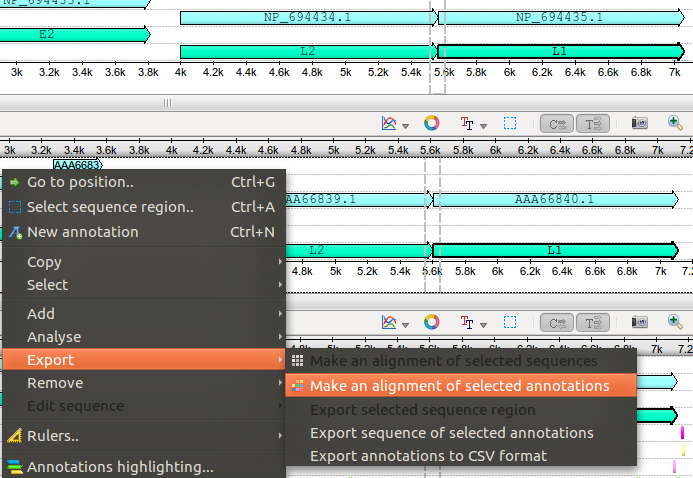
http://ugene.unipro.ru/yabbfiles/Attachments/Detail_view.pngThe "random" behavior came when I had some sequence selected in the detailed view. Even hiding this views and selecting features, I get the option for translation:
 http://ugene.unipro.ru/yabbfiles/Attachments/Selected.png
http://ugene.unipro.ru/yabbfiles/Attachments/Selected.pngThen, the problem is that in this last case, Ugene will use the selected features shown in the screen if you choose the DNA alignment, while it will use the "other" selected sequences (hidden or not) when selecting the translation option. This could be solved with an automatic selection in the detail view. I mean, automatically selecting the corresponding sequence in the detail view when selecting a feature in the zoom view. Also, for some situations It would be much faster than having to select a feature and then going to "Select/Sequence around selected annotation".
I can't think of a negative side for that behavior (actually, I believe it would be a nice feature).
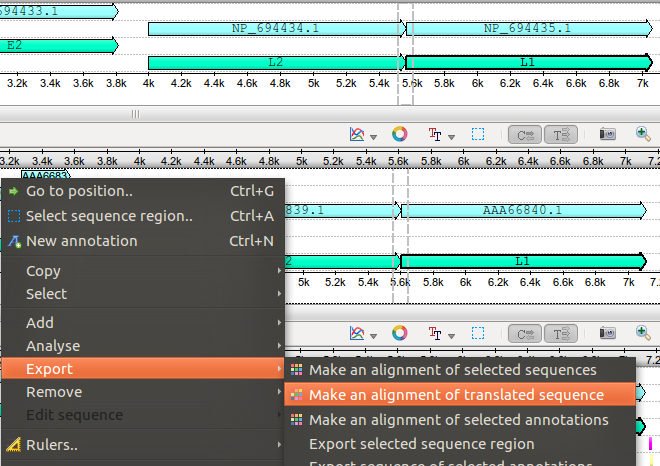
Just to make it more clear, after selecting a feature with one click it should look like this:
 http://ugene.unipro.ru/yabbfiles/Attachments/automatic.png
http://ugene.unipro.ru/yabbfiles/Attachments/automatic.pngHope this helps...
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=Detail_view.png (114 KB |
)
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=No_selection.png (94 KB |
)
 https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=
https://forum.ugene.net/forum/YaBB.pl?action=downloadfile;file=Selected.png (89 KB |
)
 pic.PNG (32 KB | )
pic.PNG (32 KB | )