Thank you very much for fixing the bug I reported a few days ago (
http://ugene.unipro.ru/forum/YaBB.pl?num=1296858562) so fast. It' s working fine now. Additionally I have two suggestions for you which would in my opinion simplify the working with the cloning pipeline.
1. Since the widely used expression vectors of the pET system (Novagen,
http://www.emdchemicals.com/life-science-research/pet/c_2tOb.s1OkacAAAEjWhl9.zLX) are numbered by the pBR322 convention (the T7 expression region is reversed on the
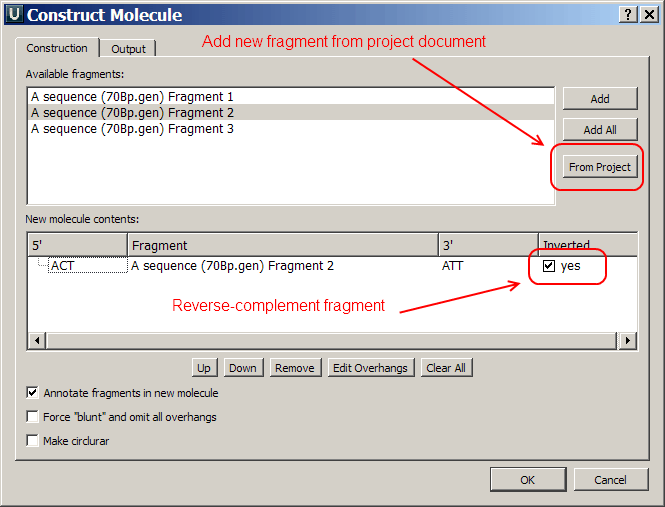
circular map) it would be nice to be able to reverse complement a fragment during cloning (without first creating a new molecule in Ugene). Maybe you could add an option like "Reverse complement Fragment" or "Invert fragment" in the "Edit molecule fragment" dialog of the cloning pipeline.
2. Since you are sometimes cloning PCR fragments into a blunt end cutted vector, it may be useful to be able to add DNA molecules from the current Ugene project into the cloning pipeline. Maybe you could add an option like "Add molecule from current project" to the "construct molecule" dialog. Due to clarity, I would not suggest to add all molecules to the "Available fragments" selection by default.
Thank you again for Ugene.
Best regards
 cloning_in_silico_new_features.gif (21 KB | )
cloning_in_silico_new_features.gif (21 KB | )